This R package combines taxonomic classifications of
high-throughput 16S rRNA gene sequences with reference
proteomes of archaea and bacteria to generate the amino acid
compositions of community reference proteomes.
Taxonomic classifications can be read from the output of the RDP
Classifier or from phyloseq-class objects created using
the Bioconductor package phyloseq.
The amino acid compositions of community reference proteomes are used to calculate chemical metrics such as carbon oxidation state (ZC) and stoichiometric hydration state (nH2O). Lower nH2O is associated with increasing salinity in samples from the Baltic Sea:
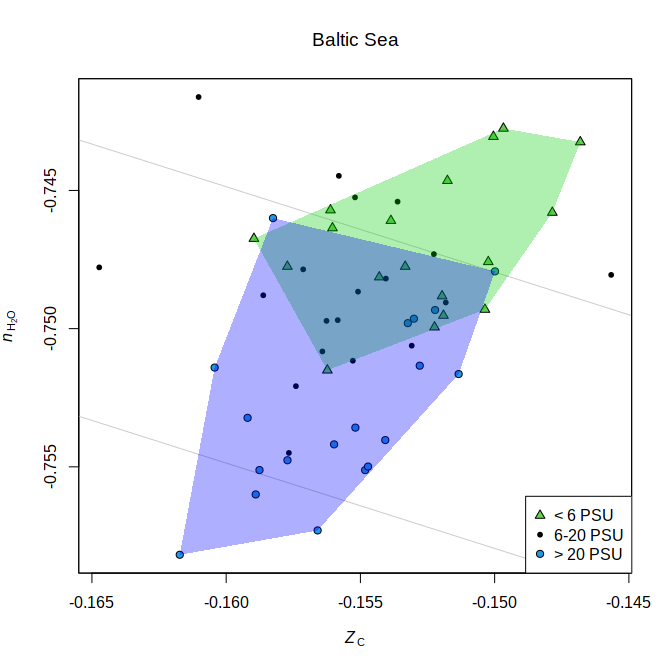
The code to make this plot is from the help page for the plot_metrics
function and uses sequence data reported by Herlemann et
al. (2016).
Precomputed amino acid compositions of reference proteomes are
provided for the Genome Taxonomy
Database (GTDB release 207) and the NCBI Reference Sequence
Database (RefSeq release 206). See the files in inst/extdata/RefSeq for the
steps used to download protein sequences from RefSeq and calculate the
total amino acid composition for each NCBI taxonomic ID (taxid). The
taxon_AA.R scripts for GTDB
and RefSeq were used to generate the
reference proteomes for genus- and higher-level archaeal and bacterial
taxa (and viruses for RefSeq).
For 16S rRNA gene sequences classified with a GTDB-based training set (see DADA2 formatted 16S rRNA gene sequences for both bacteria & archaea), taxonomic ranks and names are matched to the GTDB taxonomy to look up reference proteomes.
For 16S rRNA gene sequences classified using the default training set of the RDP Classifier, chem16S includes manual mapping to the RefSeq taxonomy as described by Dick and Tan (2023).
After installing phyloseq from Bioconductor, use
install_github (provided by either remotes
or devtools) to install chem16S from
GitHub.
# Install 'BiocManager' from CRAN
if(!require("BiocManager", quietly = TRUE)) install.packages("BiocManager")
# Install 'phyloseq' from Bioconductor
BiocManager::install("phyloseq")
# Install 'remotes' from CRAN
if(!require("remotes", quietly = TRUE)) install.packages("remotes")
# Install 'chem16S' from GitHub
remotes::install_github("jedick/chem16S", build_vignettes = TRUE)