
Visualize and tabulate single-choice, multiple-choice, matrix-style questions from survey data. Includes ability to group cross-tabulations, frequency distributions, and plots by categorical variables and to integrate survey weights. Ideal for quickly uncovering descriptive patterns in survey data.
# install.packages("devtools"), if not already downloaded
devtools::install_github("liamhaller/surveyexplorer")The data used in the following examples is from the berlinbears dataset, a fictional survey of bears in Berlin, that is included in the surveyexplorer package.
| Question: income | ||
| <1000 | ||
|---|---|---|
| 1000-2000 | ||
| 2000-3000 | ||
| 3000-4000 | ||
| 5000+ | ||
| No answer | ||
| NA | ||
| Column Total | ||
Use group_by = to partition the question into several groups
| Question: income | ||||||||
| grouped by: gender | ||||||||
| <1000 | ||||||||
|---|---|---|---|---|---|---|---|---|
| 1000-2000 | ||||||||
| 2000-3000 | ||||||||
| 3000-4000 | ||||||||
| 5000+ | ||||||||
| No answer | ||||||||
| NA | ||||||||
| Columnwise Total | ||||||||
Ignore unwanted subgroups with subgroups_to_exclude
| Question: income | ||||||
| grouped by: gender | ||||||
| <1000 | ||||||
|---|---|---|---|---|---|---|
| 1000-2000 | ||||||
| 2000-3000 | ||||||
| 3000-4000 | ||||||
| 5000+ | ||||||
| No answer | ||||||
| NA | ||||||
| Columnwise Total | ||||||
Remove NAs from the question variable with na.rm
single_table(berlinbears,
question = income,
group_by = gender,
subgroups_to_exclude = NA,
na.rm = TRUE)| Question: income | ||||||
| grouped by: gender | ||||||
| <1000 | ||||||
|---|---|---|---|---|---|---|
| 1000-2000 | ||||||
| 2000-3000 | ||||||
| 3000-4000 | ||||||
| 5000+ | ||||||
| No answer | ||||||
| Columnwise Total | ||||||
Finally, you can specify survey weights using the weight option
single_table(berlinbears,
question = income,
group_by = gender,
subgroups_to_exclude = NA,
na.rm = TRUE,
weights = weights)| Question: income | ||||||
| grouped by: gender | ||||||
| <1000 | ||||||
|---|---|---|---|---|---|---|
| 1000-2000 | ||||||
| 2000-3000 | ||||||
| 3000-4000 | ||||||
| 5000+ | ||||||
| No answer | ||||||
| Columnwise Total | ||||||
| Frequencies and counts are weighted | ||||||
The same syntax can be applied to the single_freq function to plot frequencies of the question optionally partitioned by subgroups.
single_freq(berlinbears,
question = income,
group_by = gender,
subgroups_to_exclude = NA,
na.rm = TRUE,
weights = weights)
The options and syntax for multiple-choice tables multi_table and graphs multi_graphs are the same. The only difference is the question input also accommodates tidyselect syntax to select several columns for each answer option. For example, the question “will_eat” has five answer options each prefixed by “will_eat”
berlinbears |>
dplyr::select(starts_with('will_eat')) |>
head()
#> will_eat.SQ001 will_eat.SQ002 will_eat.SQ003 will_eat.SQ004 will_eat.SQ005
#> 1 0 1 0 1 1
#> 2 0 1 1 1 1
#> 3 1 1 0 1 1
#> 4 0 0 0 1 0
#> 5 0 0 0 1 1
#> 6 0 0 0 1 0The same syntax can be used to select the question for the multiple choice tables and graphs
multi_table(berlinbears,
question = dplyr::starts_with('will_eat'),
group_by = genus,
subgroups_to_exclude = NA,
na.rm = TRUE)| Question: dplyr::starts_with(“will_eat”) | ||||||
| grouped by: genus | ||||||
| will_eat.SQ001 | ||||||
|---|---|---|---|---|---|---|
| will_eat.SQ002 | ||||||
| will_eat.SQ003 | ||||||
| will_eat.SQ004 | ||||||
| will_eat.SQ005 | ||||||
| Columnwise Total | ||||||
For graphing, the multi_freq function creates an UpSet plot to visualize the frequencies of the intersecting sets for each answer combination and also includes the ability to specify weights.
multi_freq(berlinbears,
question = dplyr::starts_with('will_eat'),
na.rm = TRUE,
weights = weights)
#> Estimes are only preciese to one significant digit, weights may have been rounded
The graphs can also be grouped
multi_freq(berlinbears,
question = dplyr::starts_with('will_eat'),
group_by = genus,
subgroups_to_exclude = NA,
na.rm = FALSE,
weights = weights)
#> Estimes are only preciese to one significant digit, weights may have been rounded
matrix_table has the same syntax as above and works with array or categorical questions
matrix_table(berlinbears,
dplyr::starts_with('p_'),
group_by = is_parent)
matrix_table(berlinbears,
dplyr::starts_with('c_'),
group_by = is_parent)| Question: dplyr::starts_with(“p_”) | ||||||
| grouped by: is_parent | ||||||
| 0 | ||||||
|---|---|---|---|---|---|---|
| p_eatstrash | ||||||
| p_hibernates | ||||||
| p_likes_zoo | ||||||
| p_likeshoney | ||||||
| p_likespine | ||||||
| p_swims | ||||||
| 1 | ||||||
| p_eatstrash | ||||||
| p_hibernates | ||||||
| p_likes_zoo | ||||||
| p_likeshoney | ||||||
| p_likespine | ||||||
| p_swims | ||||||
| Question: dplyr::starts_with(“c_”) | ||||
| grouped by: is_parent | ||||
| 0 | ||||
|---|---|---|---|---|
| c_diet | ||||
| c_exercise | ||||
| 1 | ||||
| c_diet | ||||
| c_exercise | ||||
matrix_freq visualizes the frequencies of responses


For array/matrix style questions that are numeric matrix_mean plots the mean values and confidence intervals
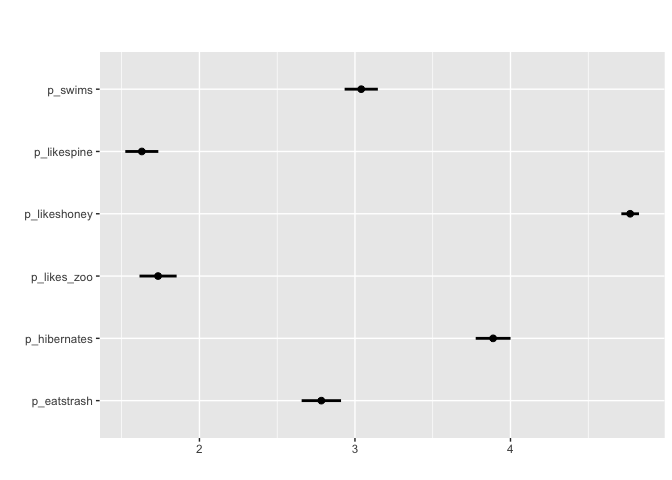
#Can also apply grouping
matrix_mean(berlinbears,
question = dplyr::starts_with('p_'),
na.rm = TRUE,
group_by = species,
subgroups_to_exclude = NA)
#with survey weights
matrix_mean(berlinbears,
question = dplyr::starts_with('p_'),
na.rm = TRUE,
group_by = species,
subgroups_to_exclude = NA,
weights = weights)
Finally, for Likert questions (scales of 3,5,7,9…) matrix_likert provides a custom plot

#you can specify custom labels with the `label` argument
matrix_likert(berlinbears,
question = dplyr::starts_with('p_'),
labels = c('Strongly disagree', 'Disagree','Neutral','Agree','Strongly agree'))
#and pass colors using the colors option
matrix_likert(berlinbears,
question = dplyr::starts_with('p_'),
labels = c('Strongly disagree', 'Disagree','Neutral','Agree','Strongly agree'),
colors = c("#E1AA28", "#1E5F46", "#7E8F75", "#EFCD83", "#E17832"))
#can also apply weights
matrix_likert(berlinbears,
question = dplyr::starts_with('p_'),
labels = c('Strongly disagree', 'Disagree','Neutral','Agree','Strongly agree'),
colors = c("#E1AA28", "#1E5F46", "#7E8F75", "#EFCD83", "#E17832"),
weights = weights) 
single_tablesingle_freqmulti_tablemulti_freqmatrix_tablematrix_freqmatrix_meanmatrix_likert*_table functions return a gt table of the cross tabulations and frequencies for each question while *_freq returns the same data but as a plot.
For matrix-style questions with numerical input, matrix_mean plots the mean value value and ± two standard deviations. matrix_likert visualizes questions that accept Likert responses (strongly agree-strongly disagree) or questions with 3,5,7,9… categories.
Each function contains the following options